Dharani Suresh
API Project 2 - Observing Vegetation Health From Space 🔬🚀
Analyzing the Cameron Peak Fire, Colorado, USA location
Brief Background: The Cameron Peak Fire was the largest fire in Colorado history, with 326 square miles burned. Looking at the destruction and recovery of vegetation using NDVI (Normalized Difference Vegetation Index). How does it work? First, we need to learn about spectral reflectance signatures.
Every object reflects some wavelengths of light more or less than others. We can see this with our eyes, since, for example, plants reflect a lot of green in the summer, and then as that green diminishes in the fall they look more yellow or orange. The image below shows spectral signatures for water, soil, and vegetation:
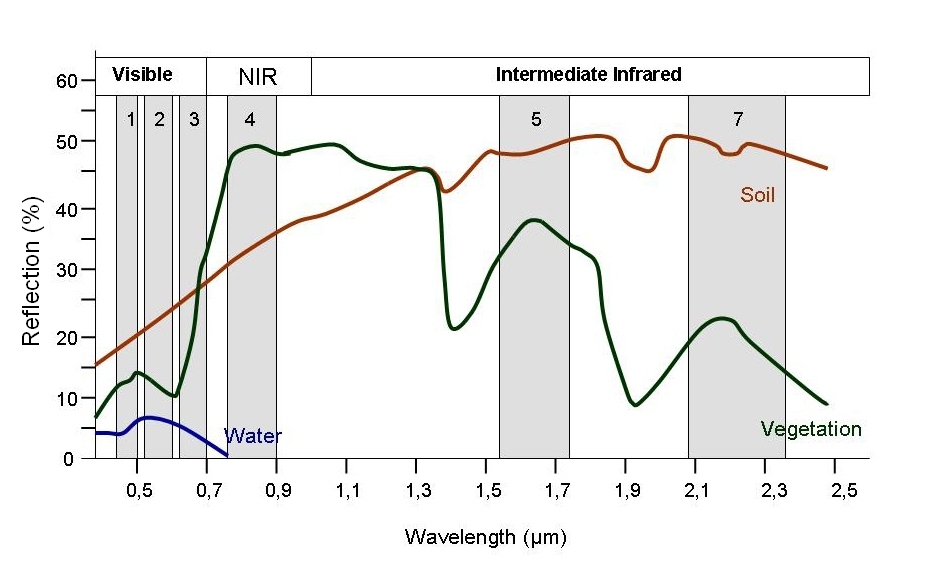 > Image source: SEOS
Project
> Image source: SEOS
Project
Healthy vegetation reflects a lot of Near-InfraRed (NIR) radiation. Dead ve Less healthy vegetation reflects a similar amounts of the visible light spectra, but less NIR radiation. We don’t see a huge drop in Green radiation until the plant is very stressed or dead. That means that NIR allows us to get ahead of what we can see with our eyes.
 > Image source: Spectral signature literature review by
px39n
> Image source: Spectral signature literature review by
px39n
Different species of plants reflect different spectral signatures, but the pattern of the signatures are similar. NDVI compares the amount of NIR reflectance to the amount of Red reflectance, thus accounting for many of the species differences and isolating the health of the plant. The formula for calculating NDVI is:
\[NDVI = \frac{(NIR - Red)}{(NIR + Red)}\]Read more about NDVI and other vegetation indices: * earthdatascience.org * USGS
Import necessary libraries
In the cell below, making sure to keep the packages in order, add packages for:
- Working with DataFrames
- Working with GeoDataFrames
What are we using the rest of these packages for? See if you can figure it out as you complete the notebook.
# Install the development version of the earthpy package
!pip install git+https://github.com/earthlab/earthpy@apppears
Collecting git+https://github.com/earthlab/earthpy@apppears
Cloning https://github.com/earthlab/earthpy (to revision apppears) to /tmp/pip-req-build-1eml1zig
Running command git clone --filter=blob:none --quiet https://github.com/earthlab/earthpy /tmp/pip-req-build-1eml1zig
Running command git checkout -b apppears --track origin/apppears
Switched to a new branch 'apppears'
Branch 'apppears' set up to track remote branch 'apppears' from 'origin'.
Resolved https://github.com/earthlab/earthpy to commit 20e0b17e563da17ba46dc32832cf4ed9173a7531
Installing build dependencies ... [?25ldone
[?25h Getting requirements to build wheel ... [?25ldone
[?25h Preparing metadata (pyproject.toml) ... [?25ldone
[?25hRequirement already satisfied: geopandas in /opt/conda/lib/python3.11/site-packages (from earthpy==0.10.0) (0.14.2)
Requirement already satisfied: matplotlib>=2.0.0 in /opt/conda/lib/python3.11/site-packages (from earthpy==0.10.0) (3.8.4)
Requirement already satisfied: numpy>=1.14.0 in /opt/conda/lib/python3.11/site-packages (from earthpy==0.10.0) (1.24.3)
Requirement already satisfied: rasterio in /opt/conda/lib/python3.11/site-packages (from earthpy==0.10.0) (1.3.9)
Requirement already satisfied: scikit-image in /opt/conda/lib/python3.11/site-packages (from earthpy==0.10.0) (0.22.0)
Requirement already satisfied: requests in /opt/conda/lib/python3.11/site-packages (from earthpy==0.10.0) (2.31.0)
Requirement already satisfied: keyring in /opt/conda/lib/python3.11/site-packages (from earthpy==0.10.0) (25.1.0)
Requirement already satisfied: contourpy>=1.0.1 in /opt/conda/lib/python3.11/site-packages (from matplotlib>=2.0.0->earthpy==0.10.0) (1.2.0)
Requirement already satisfied: cycler>=0.10 in /opt/conda/lib/python3.11/site-packages (from matplotlib>=2.0.0->earthpy==0.10.0) (0.11.0)
Requirement already satisfied: fonttools>=4.22.0 in /opt/conda/lib/python3.11/site-packages (from matplotlib>=2.0.0->earthpy==0.10.0) (4.51.0)
Requirement already satisfied: kiwisolver>=1.3.1 in /opt/conda/lib/python3.11/site-packages (from matplotlib>=2.0.0->earthpy==0.10.0) (1.4.4)
Requirement already satisfied: packaging>=20.0 in /opt/conda/lib/python3.11/site-packages (from matplotlib>=2.0.0->earthpy==0.10.0) (24.0)
Requirement already satisfied: pillow>=8 in /opt/conda/lib/python3.11/site-packages (from matplotlib>=2.0.0->earthpy==0.10.0) (10.3.0)
Requirement already satisfied: pyparsing>=2.3.1 in /opt/conda/lib/python3.11/site-packages (from matplotlib>=2.0.0->earthpy==0.10.0) (3.0.9)
Requirement already satisfied: python-dateutil>=2.7 in /opt/conda/lib/python3.11/site-packages (from matplotlib>=2.0.0->earthpy==0.10.0) (2.9.0)
Requirement already satisfied: fiona>=1.8.21 in /opt/conda/lib/python3.11/site-packages (from geopandas->earthpy==0.10.0) (1.9.6)
Requirement already satisfied: pandas>=1.4.0 in /opt/conda/lib/python3.11/site-packages (from geopandas->earthpy==0.10.0) (2.2.1)
Requirement already satisfied: pyproj>=3.3.0 in /opt/conda/lib/python3.11/site-packages (from geopandas->earthpy==0.10.0) (3.6.1)
Requirement already satisfied: shapely>=1.8.0 in /opt/conda/lib/python3.11/site-packages (from geopandas->earthpy==0.10.0) (2.0.4)
Requirement already satisfied: jaraco.classes in /opt/conda/lib/python3.11/site-packages (from keyring->earthpy==0.10.0) (3.4.0)
Requirement already satisfied: jaraco.functools in /opt/conda/lib/python3.11/site-packages (from keyring->earthpy==0.10.0) (4.0.1)
Requirement already satisfied: jaraco.context in /opt/conda/lib/python3.11/site-packages (from keyring->earthpy==0.10.0) (5.3.0)
Requirement already satisfied: importlib-metadata>=4.11.4 in /opt/conda/lib/python3.11/site-packages (from keyring->earthpy==0.10.0) (7.1.0)
Requirement already satisfied: SecretStorage>=3.2 in /opt/conda/lib/python3.11/site-packages (from keyring->earthpy==0.10.0) (3.3.3)
Requirement already satisfied: jeepney>=0.4.2 in /opt/conda/lib/python3.11/site-packages (from keyring->earthpy==0.10.0) (0.8.0)
Requirement already satisfied: affine in /opt/conda/lib/python3.11/site-packages (from rasterio->earthpy==0.10.0) (2.3.0)
Requirement already satisfied: attrs in /opt/conda/lib/python3.11/site-packages (from rasterio->earthpy==0.10.0) (23.2.0)
Requirement already satisfied: certifi in /opt/conda/lib/python3.11/site-packages (from rasterio->earthpy==0.10.0) (2024.2.2)
Requirement already satisfied: click>=4.0 in /opt/conda/lib/python3.11/site-packages (from rasterio->earthpy==0.10.0) (8.1.7)
Requirement already satisfied: cligj>=0.5 in /opt/conda/lib/python3.11/site-packages (from rasterio->earthpy==0.10.0) (0.7.2)
Requirement already satisfied: snuggs>=1.4.1 in /opt/conda/lib/python3.11/site-packages (from rasterio->earthpy==0.10.0) (1.4.7)
Requirement already satisfied: click-plugins in /opt/conda/lib/python3.11/site-packages (from rasterio->earthpy==0.10.0) (1.1.1)
Requirement already satisfied: setuptools in /opt/conda/lib/python3.11/site-packages (from rasterio->earthpy==0.10.0) (69.5.1)
Requirement already satisfied: charset-normalizer<4,>=2 in /opt/conda/lib/python3.11/site-packages (from requests->earthpy==0.10.0) (3.3.2)
Requirement already satisfied: idna<4,>=2.5 in /opt/conda/lib/python3.11/site-packages (from requests->earthpy==0.10.0) (3.7)
Requirement already satisfied: urllib3<3,>=1.21.1 in /opt/conda/lib/python3.11/site-packages (from requests->earthpy==0.10.0) (2.2.1)
Requirement already satisfied: scipy>=1.8 in /opt/conda/lib/python3.11/site-packages (from scikit-image->earthpy==0.10.0) (1.12.0)
Requirement already satisfied: networkx>=2.8 in /opt/conda/lib/python3.11/site-packages (from scikit-image->earthpy==0.10.0) (3.1)
Requirement already satisfied: imageio>=2.27 in /opt/conda/lib/python3.11/site-packages (from scikit-image->earthpy==0.10.0) (2.33.1)
Requirement already satisfied: tifffile>=2022.8.12 in /opt/conda/lib/python3.11/site-packages (from scikit-image->earthpy==0.10.0) (2023.4.12)
Requirement already satisfied: lazy_loader>=0.3 in /opt/conda/lib/python3.11/site-packages (from scikit-image->earthpy==0.10.0) (0.3)
Requirement already satisfied: six in /opt/conda/lib/python3.11/site-packages (from fiona>=1.8.21->geopandas->earthpy==0.10.0) (1.16.0)
Requirement already satisfied: zipp>=0.5 in /opt/conda/lib/python3.11/site-packages (from importlib-metadata>=4.11.4->keyring->earthpy==0.10.0) (3.17.0)
Requirement already satisfied: pytz>=2020.1 in /opt/conda/lib/python3.11/site-packages (from pandas>=1.4.0->geopandas->earthpy==0.10.0) (2024.1)
Requirement already satisfied: tzdata>=2022.7 in /opt/conda/lib/python3.11/site-packages (from pandas>=1.4.0->geopandas->earthpy==0.10.0) (2023.3)
Requirement already satisfied: cryptography>=2.0 in /opt/conda/lib/python3.11/site-packages (from SecretStorage>=3.2->keyring->earthpy==0.10.0) (42.0.5)
Requirement already satisfied: more-itertools in /opt/conda/lib/python3.11/site-packages (from jaraco.classes->keyring->earthpy==0.10.0) (10.2.0)
Requirement already satisfied: backports.tarfile in /opt/conda/lib/python3.11/site-packages (from jaraco.context->keyring->earthpy==0.10.0) (1.1.1)
Requirement already satisfied: cffi>=1.12 in /opt/conda/lib/python3.11/site-packages (from cryptography>=2.0->SecretStorage>=3.2->keyring->earthpy==0.10.0) (1.16.0)
Requirement already satisfied: pycparser in /opt/conda/lib/python3.11/site-packages (from cffi>=1.12->cryptography>=2.0->SecretStorage>=3.2->keyring->earthpy==0.10.0) (2.22)
import getpass
import json
import os
import pathlib
from glob import glob
import pandas as pd
import geopandas as gpd
import earthpy.appeears as eaapp
import hvplot.pandas
import hvplot.xarray
import rioxarray as rxr
import xarray as xr
data_dir = os.path.join(pathlib.Path.home(), 'ndvi-data')
# Make the data directory
os.makedirs(data_dir, exist_ok=True)
data_dir
'/home/jovyan/ndvi-data'
Study Area: Cameron Peak Fire Boundary
Earth Data Science data formats
In Earth Data Science, we get data in three main formats:
| Data type | Descriptions | Common file formats | Python type |
|---|---|---|---|
| Time Series | The same data points (e.g. streamflow) collected multiple times over time | Tabular formats (e.g. .csv, or .xlsx) | pandas DataFrame |
| Vector | Points, lines, and areas (with coordinates) | Shapefile (often an archive like a .zip file because a Shapefile is actually a collection of at least 3 files) |
geopandas GeoDataFrame |
| Raster | Evenly spaced spatial grid (with coordinates) | GeoTIFF (.tif), NetCDF (.nc), HDF (.hdf) |
rioxarray DataArray |
Read more
Check out the sections about about vector data and raster data in the textbook.
# Download the Cameron Peak fire boundary
cameron_url = ("https://services3.arcgis.com/T4QMspbfLg3qTGWY/arcgis/rest/services/WFIGS_Interagency_Perimeters/FeatureServer/0/query?where=poly_IncidentName%20%3D%20'CAMERON%20PEAK'&outFields=*&outSR=4326&f=json")
cameron_url
"https://services3.arcgis.com/T4QMspbfLg3qTGWY/arcgis/rest/services/WFIGS_Interagency_Perimeters/FeatureServer/0/query?where=poly_IncidentName%20%3D%20'CAMERON%20PEAK'&outFields=*&outSR=4326&f=json"
cameron_gdf = gpd.read_file(
cameron_url, index_col='DATE', parse_dates=True, na_values='NaN')
cameron_gdf
/opt/conda/lib/python3.11/site-packages/geopandas/io/file.py:399: FutureWarning: errors='ignore' is deprecated and will raise in a future version. Use to_datetime without passing `errors` and catch exceptions explicitly instead
as_dt = pd.to_datetime(df[k], errors="ignore")
/opt/conda/lib/python3.11/site-packages/geopandas/io/file.py:399: FutureWarning: errors='ignore' is deprecated and will raise in a future version. Use to_datetime without passing `errors` and catch exceptions explicitly instead
as_dt = pd.to_datetime(df[k], errors="ignore")
/opt/conda/lib/python3.11/site-packages/geopandas/io/file.py:399: FutureWarning: errors='ignore' is deprecated and will raise in a future version. Use to_datetime without passing `errors` and catch exceptions explicitly instead
as_dt = pd.to_datetime(df[k], errors="ignore")
/opt/conda/lib/python3.11/site-packages/geopandas/io/file.py:399: FutureWarning: errors='ignore' is deprecated and will raise in a future version. Use to_datetime without passing `errors` and catch exceptions explicitly instead
as_dt = pd.to_datetime(df[k], errors="ignore")
/opt/conda/lib/python3.11/site-packages/geopandas/io/file.py:399: FutureWarning: errors='ignore' is deprecated and will raise in a future version. Use to_datetime without passing `errors` and catch exceptions explicitly instead
as_dt = pd.to_datetime(df[k], errors="ignore")
/opt/conda/lib/python3.11/site-packages/geopandas/io/file.py:399: FutureWarning: errors='ignore' is deprecated and will raise in a future version. Use to_datetime without passing `errors` and catch exceptions explicitly instead
as_dt = pd.to_datetime(df[k], errors="ignore")
/opt/conda/lib/python3.11/site-packages/geopandas/io/file.py:399: FutureWarning: errors='ignore' is deprecated and will raise in a future version. Use to_datetime without passing `errors` and catch exceptions explicitly instead
as_dt = pd.to_datetime(df[k], errors="ignore")
/opt/conda/lib/python3.11/site-packages/geopandas/io/file.py:399: FutureWarning: errors='ignore' is deprecated and will raise in a future version. Use to_datetime without passing `errors` and catch exceptions explicitly instead
as_dt = pd.to_datetime(df[k], errors="ignore")
/opt/conda/lib/python3.11/site-packages/geopandas/io/file.py:399: FutureWarning: errors='ignore' is deprecated and will raise in a future version. Use to_datetime without passing `errors` and catch exceptions explicitly instead
as_dt = pd.to_datetime(df[k], errors="ignore")
/opt/conda/lib/python3.11/site-packages/geopandas/io/file.py:399: FutureWarning: errors='ignore' is deprecated and will raise in a future version. Use to_datetime without passing `errors` and catch exceptions explicitly instead
as_dt = pd.to_datetime(df[k], errors="ignore")
/opt/conda/lib/python3.11/site-packages/geopandas/io/file.py:399: FutureWarning: errors='ignore' is deprecated and will raise in a future version. Use to_datetime without passing `errors` and catch exceptions explicitly instead
as_dt = pd.to_datetime(df[k], errors="ignore")
/opt/conda/lib/python3.11/site-packages/geopandas/io/file.py:399: FutureWarning: errors='ignore' is deprecated and will raise in a future version. Use to_datetime without passing `errors` and catch exceptions explicitly instead
as_dt = pd.to_datetime(df[k], errors="ignore")
/opt/conda/lib/python3.11/site-packages/geopandas/io/file.py:399: FutureWarning: errors='ignore' is deprecated and will raise in a future version. Use to_datetime without passing `errors` and catch exceptions explicitly instead
as_dt = pd.to_datetime(df[k], errors="ignore")
/opt/conda/lib/python3.11/site-packages/geopandas/io/file.py:399: FutureWarning: errors='ignore' is deprecated and will raise in a future version. Use to_datetime without passing `errors` and catch exceptions explicitly instead
as_dt = pd.to_datetime(df[k], errors="ignore")
/opt/conda/lib/python3.11/site-packages/geopandas/io/file.py:399: FutureWarning: errors='ignore' is deprecated and will raise in a future version. Use to_datetime without passing `errors` and catch exceptions explicitly instead
as_dt = pd.to_datetime(df[k], errors="ignore")
| OBJECTID | poly_SourceOID | poly_IncidentName | poly_FeatureCategory | poly_MapMethod | poly_GISAcres | poly_CreateDate | poly_DateCurrent | poly_PolygonDateTime | poly_IRWINID | ... | attr_ModifiedOnDateTime_dt | attr_Source | attr_IsCpxChild | attr_CpxName | attr_CpxID | attr_SourceGlobalID | GlobalID | Shape__Area | Shape__Length | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 14659 | 10393 | Cameron Peak | Wildfire Final Fire Perimeter | Infrared Image | 208913.3 | NaT | 2023-03-14 15:13:42.810000+00:00 | 2020-10-06 21:30:00+00:00 | {53741A13-D269-4CD5-AF91-02E094B944DA} | ... | 2021-12-06 19:48:12.647000+00:00 | FODR | None | None | None | {5F5EC9BA-5F85-495B-8B97-1E0969A1434E} | e8e4ffd4-db84-4d47-bd32-d4b0a8381fff | 0.089957 | 5.541765 | MULTIPOLYGON (((-105.88333 40.54598, -105.8837... |
1 rows × 114 columns
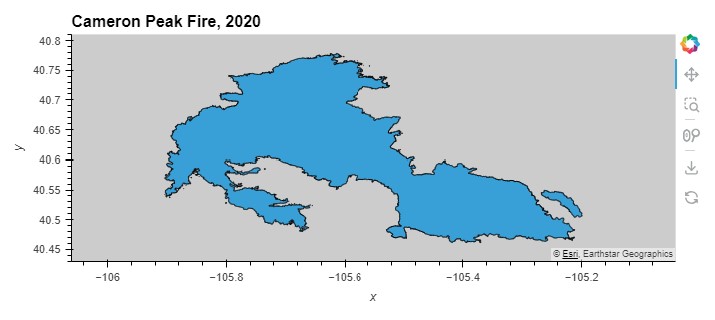
Site Map
# Plot the Cameron Peak Fire boundary
#cameron_gdf.explore()
cameron_gdf.hvplot(
title='Cameron Peak Fire, 2020',
tiles='EsriImagery')
Exploring the AppEEARS API for NASA Earthdata access
Downloading MODIS NDVI data for the study period. MODIS is a multispectral instrument that measures Red and NIR data (and so can be used for NDVI). There are two MODIS sensors on two different platforms: satellites Terra and Aqua.
Read more
Learn more about MODIS datasets and the science they support
A special download that covers only the Cameron study area is not readily available through a direct link; instead, negotiations must be conducted with the data server. This is facilitated using the APPEEARS API (Application Programming Interface). The API enables data requests to be made using code. The handling of these API requests can be managed using code from the earthpy library.
# Initialize AppeearsDownloader for MODIS NDVI data
ndvi_downloader = eaapp.AppeearsDownloader(
download_key='cp-ndvi',
ea_dir=data_dir,
product='MOD13Q1.061',
layer='_250m_16_days_NDVI',
start_date="07-01",
end_date="07-31",
recurring=True,
year_range=[2018, 2023],
polygon=cameron_gdf
)
ndvi_downloader.download_files(cache=True)
Putting it together: Working with multi-file raster datasets in Python
All downloaded files need to be loaded into Python, beginning with the retrieval of all the file names. Additionally, the date will need to be extracted from each filename. For guidance on extracting information from filenames, refer to the lesson on getting information from filenames in the textbook.
GOTCHA ALERT
globdoesn’t necessarily find files in the order you would expect. Make sure to sort your file names like it says in the textbook.
# Get list of NDVI tif file paths
ndvi_paths = sorted(glob(os.path.join(data_dir, 'cp-ndvi', '*', '*NDVI*.tif')))
len(ndvi_paths)
18
scale_factor = 10000
doy_start = -19
doy_end = -12
ndvi_das = []
for ndvi_path in ndvi_paths:
# Get date from file name
doy = ndvi_path[doy_start:doy_end]
date = pd.to_datetime(doy, format='%Y%j')
# Open dataset
da = rxr.open_rasterio(ndvi_path, masked=True).squeeze()
# Add date dimension and clean up metadata
da = da.assign_coords({'date': date})
da = da.expand_dims({'date': 1})
da.name = 'NDVI'
# Multiple by scale factor
da = da / scale_factor
# Prepare for concatenation
ndvi_das.append(da)
len(ndvi_das)
18
Next, stacking arrays by date into a time series using the
xr.combine_by_coords() function with date dimension.
ndvi_da = xr.combine_by_coords(ndvi_das, coords=['date'])
ndvi_da
<xarray.Dataset>
Dimensions: (x: 339, y: 153, date: 18)
Coordinates:
band int64 1
* x (x) float64 -105.9 -105.9 -105.9 ... -105.2 -105.2 -105.2
* y (y) float64 40.78 40.78 40.77 40.77 ... 40.47 40.47 40.46 40.46
spatial_ref int64 0
* date (date) datetime64[ns] 2018-06-26 2018-07-12 ... 2023-07-28
Data variables:
NDVI (date, y, x) float32 0.5672 0.5711 0.6027 ... 0.6327 0.6327Plot the change in NDVI spatially
* Selecting data from 2021 to 2023 (3 years after the fire) * Taking the temporal mean (over the date, not spatially) * Getting the NDVI variable (should be a DataArray, not a Dataset) * Repeating for the data from 2018 to 2020 (3 years before the fire) * Subtracting the 2018-2020 time period from the 2021-2023 time period * Plotting the result using a diverging color map like
cmap=plt.cm.PiYGThere are different types of color maps for different types of data. In this case, we want decreases to be a different color from increases, so we should use a diverging color map. Check out available colormaps in the matplotlib documentation.
Adding the fire boundary to the plot
ndvi_diff = (
ndvi_da
.sel(date=slice('2021', '2023'))
.mean('date')
.NDVI
- ndvi_da
.sel(date=slice('2018', '2020'))
.mean('date')
.NDVI
)
(
ndvi_diff.hvplot(x='x', y='y', cmap='PiYG', geo=True)
*
cameron_gdf.hvplot(geo=True, fill_color=None, line_color='black')
)
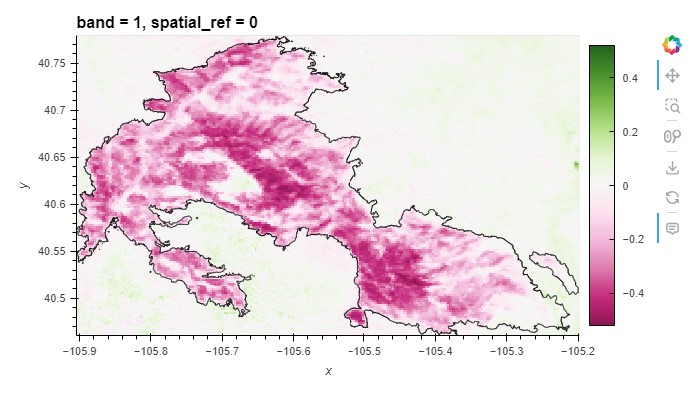
Is the NDVI lower within the fire boundary after the fire?
Computing the mean NDVI inside and outside the fire boundary.
Using the code below to get a GeoDataFrame of the area outside
the Reservation. * Checking the variable names - making sure that
the code uses your boundary GeoDataFrame * Testing if the
geometry was modified correctly by adding some code to take a look at the
results.
out_gdf = (
gpd.GeoDataFrame(geometry=cameron_gdf.envelope)
.overlay(cameron_gdf, how='difference'))
Clipping the DataArray to the boundaries for both inside and outside
the reservation. Replacing the GeoDataFrame name with
your own. Check out the lesson on clipping data with the rioxarray
library in the
textbook.
GOTCHA ALERT
It’s important to use
from_disk=Truewhen clipping large arrays like this. It allows the computer to use less valuable memory resources when clipping - you will probably find that otherwise the cell below crashes the kernel
# Clip data to both inside and outside the boundary
ndvi_cp_da = ndvi_da.rio.clip(cameron_gdf.geometry, from_disk=True)
ndvi_out_da = ndvi_da.rio.clip(out_gdf.geometry, from_disk=True)
**
For both inside and outside the fire boundary:
- Groupping the data by year
- Taking the mean. Always need to tell reducing methods in
xarraywhat dimensions you want to reduce. Summarizing data across all dimensions, by using the...syntax, e.g..mean(...)as a shorthand.- Selecting the NDVI variable
- Converting to a DataFrame using the
to_dataframe()method- Joining the two DataFrames for plotting using the
.join()method. Renaming the columns using thelsuffix=andrsuffix=parameters
GOTCHA ALERT
The DateIndex in pandas is a little different from the Datetime Dimension in xarray. You will need to use the
.dt.yearsyntax to access information about the year, not just.year.
Finally, plotting annual July means for both inside and outside the Reservation on the same plot.
:::
# Compute mean annual July NDVI
jul_ndvi_cp_df = (
ndvi_cp_da
.groupby(ndvi_cp_da.date.dt.year)
.mean(...)
.NDVI.to_dataframe())
jul_ndvi_out_df = (
ndvi_out_da
.groupby(ndvi_out_da.date.dt.year)
.mean(...)
.NDVI.to_dataframe())
# Plot inside and outside the reservation
jul_ndvi_df = (
jul_ndvi_cp_df[['NDVI']]
.join(
jul_ndvi_out_df[['NDVI']],
lsuffix=' Burned Area', rsuffix=' Unburned Area')
)
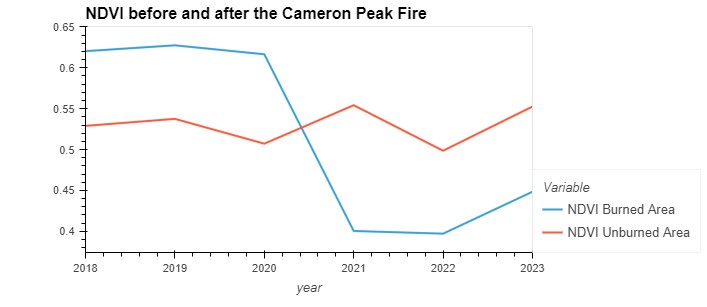
jul_ndvi_df.hvplot(
title='NDVI before and after the Cameron Peak Fire'
)
Taking the difference between outside and inside the Reservation and plot that. What do you observe? Don’t forget to write a headline and description of your plot!
# Plot difference inside and outside the reservation
jul_ndvi_df['difference'] = (
jul_ndvi_df['NDVI Burned Area']
- jul_ndvi_df['NDVI Unburned Area'])
jul_ndvi_df.difference.hvplot(
title='Difference between NDVI within and outside the Cameron Peak Fire'
)
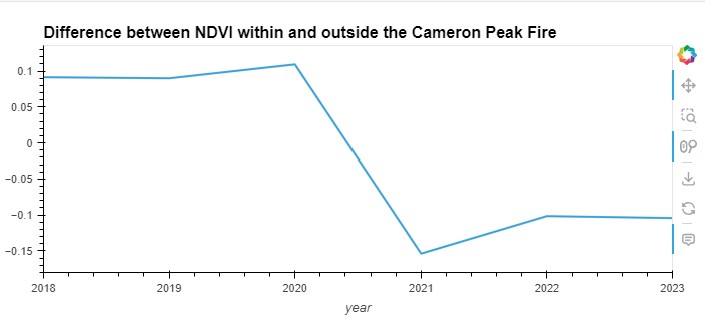
Observations from the Plot:
Sharp Decline in Burned Area:
- Year 2020: There’s a sharp decline in NDVI values in the burned area, indicating a significant reduction in vegetation due to the fire. This drop is visually prominent, illustrating the immediate impact of the fire on the vegetation.
Gradual Recovery:
- Post-2020: The NDVI in the burned area begins to recover, though it does not return to pre-fire levels by 2023. This upward trend suggests some vegetative regrowth or recovery in the burned region over the three years following the fire.
Stability in Unburned Area:
- 2018 to 2023: The NDVI values in the unburned area show some fluctuation but remain relatively stable across the years. This stability contrasts with the dramatic fluctuations in the burned area, emphasizing the impact of the fire on vegetation health.
Comparison of Trends:
- The contrasting trends between the burned and unburned areas highlight the resilience and the differential response of vegetation to fire disturbance. The unburned area serves as a reference or control, showing what the NDVI trend might have looked like absent the fire.
This plot is useful for visualizing the direct effects of wildfires on vegetation over time and can be critical for ecological studies and restoration efforts.
%%capture
%%bash
jupyter nbconvert "vegetation.ipynb" --to markdown